We study a procedure for generating a random sequence of permutations of [N]. We start with the identity permutation, and then in each step, we choose two elements uniformly at random, and swap them. We obtain a sequence of permutations, where each term is obtained from the previous one by multiplying by a uniformly-chosen transposition.
Some more formality and some technical remarks:
- This is a Markov chain, and as often with Markov chains, it would be better it was aperiodic. As described, the cycle will alternate between odd and even permutations. So we allow the two elements chosen to be the same. This laziness slows down the chain by a factor N-1/N, but removes periodicity. We will work over timescales where this adjustment makes no practical difference.
- Let
 be the sequence of transpositions. We could define the sequence of permutations by
be the sequence of transpositions. We could define the sequence of permutations by  . I find it slightly more helpful to think of swapping the elements in places i and j, rather the elements i and j themselves, and so I’ll use this language, for which
. I find it slightly more helpful to think of swapping the elements in places i and j, rather the elements i and j themselves, and so I’ll use this language, for which  is the appropriate description. Of course, transpositions and the identity are self-inverse permutations, so it makes no difference to anything we might discuss.
is the appropriate description. Of course, transpositions and the identity are self-inverse permutations, so it makes no difference to anything we might discuss.
- You can view this as lazy random walk on the Cayley graph of
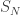 generated by the set of transpositions. That is, the vertices of the graph are elements of
generated by the set of transpositions. That is, the vertices of the graph are elements of 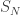 , and two are connected by an edge if one can be obtained from the other by multiplying by a transposition. Note this relation is symmetric. Hence random transposition random walk.
, and two are connected by an edge if one can be obtained from the other by multiplying by a transposition. Note this relation is symmetric. Hence random transposition random walk.
- Almost everything under discussion would work in continuous time too.
At a very general level, this sort of model is interesting because sometimes the only practical way to introduce ‘global randomness’ is repeatedly to apply ‘local randomness’. This is not the case for permutations – it is not hard to sample uniformly from 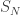 . But it is a tractable model in which to study relevant questions about the generating randomness on a complicated set through iterated local operations.
. But it is a tractable model in which to study relevant questions about the generating randomness on a complicated set through iterated local operations.
Since it is a Markov chain with a straightforward invariant distribution, we can ask about the mixing time. That is, the correct scaling for the number of moves before the random permutation is close in distribution (say in the sense of total variation distance) to the equilibrium distribution. See this series of posts for an odd collection of background material on the topic. Diaconis and Shahshahani [DS81] give an analytic argument for mixing around  transpositions. Indeed they include a constant because it is a sharp cutoff, where the total variation distance drops from approximately 1 to approximately 0 in O(N) steps.
transpositions. Indeed they include a constant because it is a sharp cutoff, where the total variation distance drops from approximately 1 to approximately 0 in O(N) steps.
Comparison with Erdos-Renyi random graph process
In the previous result, one might observe that  is also the threshold number of edges to guarantee connectivity of the Erdos-Renyi random graph G(N,m) with high probability. [ER59] Indeed, there is also a sharp transition around this threshold in this setting too.
is also the threshold number of edges to guarantee connectivity of the Erdos-Renyi random graph G(N,m) with high probability. [ER59] Indeed, there is also a sharp transition around this threshold in this setting too.
We explore this link further. We can construct a sequence of random graphs simultaneously with the random transposition random walk. When we multiply by transposition (i j), we add edge ij in the graph. Laziness of RTRW and the possibility of multiple edges mean this definition isn’t literally the same as the conventional definition of a discrete-time Erdos-Renyi random graph process, but again this is not a problem for any of the effects we seek to study.
The similarity between the constructions is clear. But what about the differences? For the RTRW, we need to track more information than the random graph. That is, we need to know what order the transpositions were added, rather than merely which edges were added. However, the trade-off is that a permutation is a simpler object than a graph in the following sense. A permutation can be a described as a union of disjoint cycles. In an exchangeable setting, all the information about a random permutation is encoded in the lengths of the these cycles. Whereas in a graph, geometry is important. It’s an elegant property of the Erdos-Renyi process that we can forget about the geometry and treat it as a process on component sizes (indeed, a multiplicative coalescent process), but there are other questions we might need to ask for which we do have to study the graph structure itself.
Within this analogy, unfortunately the word cycle means different things in the two different settings. In a permutation, a cycle is a directed orbit, while in a graph it has the usual definition. I’m going to write graph-cycle whenever relevant to avoid confusion.
A first observation is that, under this equivalence, the cycles of the permutation form a finer partition than the components of the graph. This is obvious. If we split the vertices into sets A and B, and there are no edges between them, then nothing in set A will ever get moved out of set A by a transposition. (Note that the slickness of this analogy is the advantage of viewing a transposition as swapping the elements in places i and j.)
However, we might then ask under what circumstances is a cycle of the permutation the same as a component of the graph (rather than a strict subset of it). A first answer is the following:
Lemma: [Den59] The permutation formed by a product of transpositions corresponding in any order to a tree in the graph has a single cycle.
We can treat this as a standalone problem and argue in the following predictable fashion. (Indeed, I was tempted to set this as a problem during selection for the UK team for IMO 2017 – it’s perfectly suitable in this context I think.) The first transposition corresponds to some edge say ab, and removing this edge divides the vertices into components  . Since no further transposition swaps between places in A and places in B, the final permutation maps a into B and b into A, and otherwise preserves A and B.
. Since no further transposition swaps between places in A and places in B, the final permutation maps a into B and b into A, and otherwise preserves A and B.
This argument extends to later transpositions too. Now, suppose there are multiple cycles. Colour one of them. So during the process, the coloured labels move around. At some point, we must swap a coloured label with an uncoloured label. Consider this edge, between places a and b as before, and indeed the same conclusion holds. WLOG we move the coloured label from a to b. But then at the end of the process (ie in the permutation) there are more coloured labels in B than initially. But the number of coloured labels should be the same, because they just cycle around in the final permutation.
We can learn a bit more by trying thinking about the action on cycles (in the permutation) of adding a transposition. In the following pair of diagrams, the black arrows represent the original permutation (note it’s not helpful to think of the directed edges as having anything to do with transpositions now), the dashed line represents a new transposition, and the new arrows describe the new permutation which results from this product.
It’s clear from this that adding a transposition between places corresponding to different cycles causes the cycles to merge, while adding a transposition between places already in the same cycle causes the cycle to split into two cycles. Furthermore the sizes of the two cycles formed is related to the distance in the cycle between the places defining the transposition.
This allows us to prove the lemma by adding the edges of the tree one-at-a-time and using induction. The inductive claim is that cycles of the permutation exactly correspond to components of the partially-built tree. Assuming this claim guarantees that the next step is definitely a merge, not a split (otherwise the edge corresponding to the next step would have to form a cycle). If all N-1 steps are merges, then the number of cycles is reduced by one on each step, and so the final permutation must be a single cycle.
Uniform split-merge
This gives another framework for thinking about the RTRW itself, entirely in terms of cycle lengths as a partition of [N]. That is, given a partition, we choose a pair of parts in a size-biased way. If they are different, we merge them; and if it is the same part, with size k, we split them into two parts, with sizes chosen uniformly from { (1,k-1), (2,k-2), … (k-1,1) }.
What’s nice about this is that it’s easy to generalise to real-valued partitions, eg of [0,1]. Given a partition of [0,1], we sample two IID U[0,1] random variables  . If these correspond to different parts, we replace these parts by a single part with size given by the sum. If these correspond to the same part, with size
. If these correspond to different parts, we replace these parts by a single part with size given by the sum. If these correspond to the same part, with size 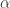 , we split this part into two parts with sizes
, we split this part into two parts with sizes  and
and  . This is equivalent in a distributional sense to sampling another U[0,1] variable U and replacing
. This is equivalent in a distributional sense to sampling another U[0,1] variable U and replacing 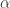 with
with  . We probably want our partition to live in
. We probably want our partition to live in 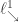 , so we might have to reorder the parts afterwards too.
, so we might have to reorder the parts afterwards too.
These uniform split-merge dynamics have a (unique) stationary distribution, the canonical Poisson-Dirichlet random partition, hereafter PD(0,1). This was first shown in [DMZZ04], and then in a framework more relevant to this post by Schramm [Sch08].
Conveniently, PD(0,1) is also the scaling limit of the cycle lengths in a uniform random permutation (scaled by N). The best way to see this is to start with the observation that the length of the cycle containing 1 in a permutation chosen uniformly from 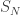 has the uniform distribution on {1,…,N}. This matches up well with the uniform stick-breaking construction of PD(0,1), though other arguments are available too. Excellent background on Poisson-Dirichlet distributions and this construction and equivalence can be found in Chapter 3 of Pitman’s comprehensive St. Flour notes [CSP]. Also see this post, and the links within, with the caveat that my understanding of the topic was somewhat shaky then (as presently, for now).
has the uniform distribution on {1,…,N}. This matches up well with the uniform stick-breaking construction of PD(0,1), though other arguments are available too. Excellent background on Poisson-Dirichlet distributions and this construction and equivalence can be found in Chapter 3 of Pitman’s comprehensive St. Flour notes [CSP]. Also see this post, and the links within, with the caveat that my understanding of the topic was somewhat shaky then (as presently, for now).
However, Schramm says slightly more than this. As the Erdos-Renyi graph passes criticality, there is a well-defined (and whp unique) giant component including 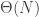 vertices. It’s not clear that the corresponding permutation should have giant cycles. Indeed, whp the giant component has
vertices. It’s not clear that the corresponding permutation should have giant cycles. Indeed, whp the giant component has 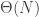 surplus edges, so the process of cycle lengths will have undergone
surplus edges, so the process of cycle lengths will have undergone  splits. Schramm shows that most of the labels within the giant component are contained in giant cycles in the permutation. Furthermore, the distribution of cycle lengths within the giant component, rescaled by the size of the giant component, converges in distribution to PD(0,1) at any supercritical time
splits. Schramm shows that most of the labels within the giant component are contained in giant cycles in the permutation. Furthermore, the distribution of cycle lengths within the giant component, rescaled by the size of the giant component, converges in distribution to PD(0,1) at any supercritical time 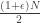
This is definitely surprising, since we already know that the whole permutation doesn’t look close to uniform until time  . Essentially, even though the size of the giant component is non-constant (ie it’s gaining vertices), the uniform split-merge process is happening to the cycles within it at rate N. So heuristically, at the level of the largest cycles, at any supercritical time we have a non-trivial partition, so at any slightly later time (eg
. Essentially, even though the size of the giant component is non-constant (ie it’s gaining vertices), the uniform split-merge process is happening to the cycles within it at rate N. So heuristically, at the level of the largest cycles, at any supercritical time we have a non-trivial partition, so at any slightly later time (eg  and
and 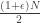 ), mixing will have comfortably occurred, and so the distribution is close to PD(0,1).
), mixing will have comfortably occurred, and so the distribution is close to PD(0,1).
This is explained very clearly in the introduction of [Ber10], in which the approach is extended to a random walk on 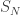 driven by a uniform choice from any conjugacy class.
driven by a uniform choice from any conjugacy class.
So this really does tell us how the global uniform randomness emerges. As the random graph process passes criticality, we have a positive mass of labels in a collection of giant cycles which are effectively a continuous-space uniform split-merge model near equilibrium (and thus with PD(0,1) marginals). The remaining cycles are small, corresponding to small trees which make up the remaining (subcritical by duality) components of the ER graph. These cycles slowly get absorbed into the giant cycles, but on a sufficiently slow timescale relevant to the split-merge dynamics that we do not need to think of a separate split-merge-with-immigration model. Total variation distance on permutations does feel the final few fixed points (corresponding to isolated vertices in the graph), hence the sharp cutoff corresponding to sharp transition in the number of isolated vertices.
References
[Ber10] – N. Berestycki – Emergence of giant cycles and slowdown transition in random transpositions and k-cycles. [arXiv version]
[CSP] – Pitman – Combinatorial stochastic processes. [pdf available]
[Den59] – Denes – the representation of a permutation as a product of a minimal number of transpositions, and its connection with the theory of graphs
[DS81] – Diaconis, Shahshahani – Generating a random permutation with random transpositions
[DMZZ04] – Diaconis, Mayer-Wolf, Zeitouni, Zerner – The Poisson-Dirichlet distribution is the unique invariant distribution for uniform split-merge transformations [link]
[ER59] – Erdos, Renyi – On random graphs I.
[Sch08] – Schramm – Compositions of random transpositions [book link]
is fixed. A graph with k types is a graph G=(V,E) together with a type function
. We will refer to a
symmetric matrix with non-negative entries as a kernel.
and a vector
satisfying
, and
a kernel, we define the inhomogeneous random graph
with k types as:
vertices have type i.
(for
) is present, independently, with probability
have type 1,
have type 2, etc etc. This makes no difference except in terms of the notation we have to use if we want to use exchangeability arguments later.
on [k], and assigns the types of the vertices of [n] in an IID fashion according to
. Essentially all the same results hold for these two models. (For example, this model with ‘random types’ can be studied by quenching the number of each type!) Often one works with whichever model seems easier for a given proof.
. The exponential form has a more natural interpretation if we ever need to turn the IRGs into a process. Additionally, it avoids the requirement to treat small values of n (for which, a priori,
might be greater than 1) separately.
.
. Obviously, one can set up an inhomogeneous random graph in a dense regime by an identical argument.
limit. But in the dense setting, one can achieve this with high confidence. When studying such statistical questions, these IRGs are often referred to as stochastic block models, and the recent survey of Abbe [Abbe] gives a very rich history of this type of problem in this setting.
for which
, where
satisfies
.
be a uniformly-chosen vertex in [n]. Clearly
, with the immediate mild notation abuse of viewing
as a probability distribution on [k].
:
, the number of type j neighbours of
is distributed as
.
is distributed as
.
, and similarly in the case j=i, so in both cases, the number of neighbours of type j is distributed approximately as
.



