A long time ago, I wrote quite a few a things about uniform trees. That is, a uniform choice from the  unrooted trees with vertex set [n]. This enumeration, normally called Cayley’s formula, has several elegant arguments, including the classical Prufer bijection. But making a uniform choice from a large set is awkward, and so we seek more probabilistic methods to sample such a tree, which might also give insight into the structure of a ‘typical’ uniform tree.
unrooted trees with vertex set [n]. This enumeration, normally called Cayley’s formula, has several elegant arguments, including the classical Prufer bijection. But making a uniform choice from a large set is awkward, and so we seek more probabilistic methods to sample such a tree, which might also give insight into the structure of a ‘typical’ uniform tree.
In another historic post, I talked about the Aldous-Broder algorithm. Here’s a quick summary. We run a random walk on the complete graph 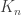 started from a uniformly-chosen vertex. Every time we arrive at a vertex we haven’t visited before, we record the edge just traversed. Eventually we have visited all n vertices, so have recorded n-1 edges. It’s easy enough to convince yourself that these n-1 edges form a tree (how could there be a cycle?) and a bit more complicated to decide that the distribution of this tree is uniform.
started from a uniformly-chosen vertex. Every time we arrive at a vertex we haven’t visited before, we record the edge just traversed. Eventually we have visited all n vertices, so have recorded n-1 edges. It’s easy enough to convince yourself that these n-1 edges form a tree (how could there be a cycle?) and a bit more complicated to decide that the distribution of this tree is uniform.
It’s worth noting that this algorithm works to construct a uniform spanning tree on any connected base graph.
This post is about a few alternative constructions and interpretations of the uniform random tree. The first construction uses a Galton-Watson process. We take a Galton-Watson process where the offspring distribution is Poisson(1), and condition that the total population size is n. The resulting random tree has a root but no labels, however if we assign labels in [n] uniformly at random, the resulting rooted tree has the uniform distribution among rooted trees on [n].
Proof
This is all about moving from ordered trees to non-ordered trees. That is, when setting up a Galton-Watson tree, we distinguish between the following two trees, drawn extremely roughly in Paint:

That is, it matters which of the first-generation vertices have three children. Anyway, for such a (rooted) ordered tree T with n vertices, the probability that the Galton-Watson process ends up equal to T is

where  is the number of children of a vertex
is the number of children of a vertex  . Then, since
. Then, since 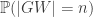 is a function of n, we find
is a function of n, we find
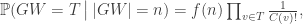
where f(n) is a function of n alone (ie depends on T only through its size n).
But given an unordered rooted tree t, labelled by [n], there are  ordered trees associated to t in the natural way. Furthermore, if we take the Poisson Galton-Watson tree conditioned to have total population size n, and label uniformly at random with [n], we obtain any one of these ordered trees with probability
ordered trees associated to t in the natural way. Furthermore, if we take the Poisson Galton-Watson tree conditioned to have total population size n, and label uniformly at random with [n], we obtain any one of these ordered trees with probability  . So the probability that we have t after we forget about the ordering is
. So the probability that we have t after we forget about the ordering is  , which is a function of n alone, and so the distribution is uniform among the set of rooted unordered trees labelled by [n], exactly as required.
, which is a function of n alone, and so the distribution is uniform among the set of rooted unordered trees labelled by [n], exactly as required.
Heuristic for Poisson offspring distribution
In this proof, the fact that 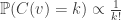 exactly balances the number of orderings of the k children explains why Poisson(1) works out. Indeed, you can see in the proof that Poisson(c) works equally well, though when
exactly balances the number of orderings of the k children explains why Poisson(1) works out. Indeed, you can see in the proof that Poisson(c) works equally well, though when  , the event we are conditioning on (namely that the total population size is n) has probability decaying exponentially in n, whereas for c=1, the branching process is critical, and the probability decays polynomially.
, the event we are conditioning on (namely that the total population size is n) has probability decaying exponentially in n, whereas for c=1, the branching process is critical, and the probability decays polynomially.
We can provide independent motivation though, from the Aldous-Broder construction. Both the conditioned Galton-Watson construction and the A-B algorithm supply the tree with a root, so we’ll keep that, and look at the distribution of the degree of the root as constructed by A-B. Let  be the vertices [n], ordered by their discovery during the construction. Then
be the vertices [n], ordered by their discovery during the construction. Then 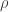 is definitely connected by an edge to
is definitely connected by an edge to 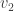 , but thereafter it follows by an elementary check that the probability
, but thereafter it follows by an elementary check that the probability 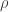 is connected to
is connected to 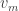 is
is  , independently across all m. In other words, the distribution of the degree of
, independently across all m. In other words, the distribution of the degree of 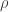 in the tree as constructed by A-B is
in the tree as constructed by A-B is
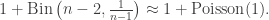
Now, in the Galton-Watson process, conditioning the tree to have fixed, large size changes the offspring distribution of the root. Conveniently though, in a limiting sense it’s the same change as conditioning the tree to have size at least n. Since these events are monotone in n, it’s possible to take a limit of the conditioning events, and interpret the result as the Galton-Watson tree conditioned to survive. It’s a beautiful result that this interpretation can be formalised as a local limit. The limiting spine decomposition consists of an infinite spine, where the offspring distribution is a size-biased version of the original offspring distribution (and so in particular, always has at least one child) and where non-spine vertices have the original distribution.
In particular, the number of the offspring of the root is size-biased, and it is well-known and not hard to check that size-biasing Poisson(c) gives 1+Poisson(c) ! So in fact we have, in an appropriate limiting sense in both objects, a match between the degree distribution of the root in the uniform tree, and in the conditioned Galton-Watson tree.
This isn’t supposed to justify why a conditioned Galton-Watson tree is relevant a priori (especially the unconditional independence of degrees), but it does explain why Poisson offspring distributions are relevant.
Construction via G(N,p) and the random cluster model
The main reason uniform trees were important to my thesis was their appearance in the Erdos-Renyi random graph G(N,p). The probability that vertices {1, …, n} form a tree component in G(N,p) with some particular structure is

Here, the first two terms give the probability that the graph structure on {1, …, n} is correct, and the the final term gives the probability of the (independent) event that these vertices are not connected to anything else in the graph. In particular, this has no dependence on the tree structure chosen on [n] (for example, whether it should be a path or a star – both examples of trees). So the conditional distribution is uniform among all trees.
If we work in some limiting regime, where  (for example if n is fixed and
(for example if n is fixed and  ), then we can get away asymptotically with less strong conditioning. Suppose we condition instead just that [n] form a component. Now, there are more ways to form a connected graph with one cycle on [n] than there are trees on [n], but the former all require an extra edge, and so the probability that a given one such tree-with-extra-edge appears as the restriction to [n] in G(N,p) is asymptotically negligible compared to the probability that the restriction to [n] of G(N,p) is a tree. Naturally, the local limit of components in G(N,c/N) is a Poisson(c) Galton-Watson branching process, and so this is all consistent with the original construction.
), then we can get away asymptotically with less strong conditioning. Suppose we condition instead just that [n] form a component. Now, there are more ways to form a connected graph with one cycle on [n] than there are trees on [n], but the former all require an extra edge, and so the probability that a given one such tree-with-extra-edge appears as the restriction to [n] in G(N,p) is asymptotically negligible compared to the probability that the restriction to [n] of G(N,p) is a tree. Naturally, the local limit of components in G(N,c/N) is a Poisson(c) Galton-Watson branching process, and so this is all consistent with the original construction.
One slightly unsatisfying aspect to this construction is that we have to embed the tree of size [n] within a much larger graph on [N] to see uniform trees. We can’t choose a scaling p=p(n) such that G(n,p) itself concentrates on trees. To guarantee connectivity with high probability, we need to take  , but by this threshold, the graph has (many) cycles with high probability.
, but by this threshold, the graph has (many) cycles with high probability.
At this PIMS summer school in Vancouver, one of the courses is focusing on lattice spin models, including the random cluster model, which we now briefly define. We start with some underlying graph G. From a physical motivation, we might take G to be 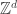 or some finite subset of it, or a d-ary tree, or the complete graph
or some finite subset of it, or a d-ary tree, or the complete graph 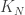 . As in classical bond percolation (note G(N,p) is bond percolation on
. As in classical bond percolation (note G(N,p) is bond percolation on 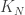 ), a random subset of the edges of G are included, or declared open. The probability of a given configuration w, with e open edges is proportional to
), a random subset of the edges of G are included, or declared open. The probability of a given configuration w, with e open edges is proportional to
 (*)
(*)
where the edge-weight 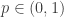 as usual, and cluster weight
as usual, and cluster weight  , and
, and  counts the number of connected components in configuration w. When q=1, we recover classical bond percolation (including G(N,p) ), while for q>1, this cluster-reweighting favours having more components, and q<1 favours fewer components. Note that in the case
counts the number of connected components in configuration w. When q=1, we recover classical bond percolation (including G(N,p) ), while for q>1, this cluster-reweighting favours having more components, and q<1 favours fewer components. Note that in the case  , the normalising constant (or partition function) of (*) is generally intractable to calculate explicitly.
, the normalising constant (or partition function) of (*) is generally intractable to calculate explicitly.
As in the Erdos-Renyi graph, consider fixing the underlying graph G, and taking 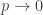 , but also taking
, but also taking  . So the resulting graph asymptotically ‘wants to have as few edges as possible, but really wants to have as few components as possible’. In particular, 1) all spanning trees of G are equally likely; 2) any configuration with more than one component has asymptotically negligible probability relative to any tree; 3) any graph with a cycle has #components + #edges greater than that of a tree, and so is asymptotically negligible probability relative to any tree.
. So the resulting graph asymptotically ‘wants to have as few edges as possible, but really wants to have as few components as possible’. In particular, 1) all spanning trees of G are equally likely; 2) any configuration with more than one component has asymptotically negligible probability relative to any tree; 3) any graph with a cycle has #components + #edges greater than that of a tree, and so is asymptotically negligible probability relative to any tree.
In other words, the limit of the distribution is the uniform spanning tree of G, and so this (like Aldous-Broder) is a substantial generalisation, which constructs the uniform random tree in the special case where  .
.
. We’ve also taken a lengthy detour to revise Galton-Watson trees, with a particular focus on the case of Poisson offspring distribution.
has some number of neighbours distributed as
, and the same approximation remains valid as we explore the graph (for example in a breadth-first fashion) either until we have seen a large number of vertices, or unless some ultra-pathological event happens, such as a vertex having degree n/3.
is well-approximated by the Galton-Watson tree with
offspring, and in this lecture and the next we try to make this notion precise, and discuss some consequences when we can show that this form of convergence occurs.
, where the vertex set of
is [n], or certainly increasing in n (as in the first example).
, we say such a sequence
converges to
locally if for all radii
, we have
. In words, the neighbourhood around
in
is the same up to radius r as the neighbourhood around
in
, so long as n is large enough (for given r).
, the binary tree to depth n.
to be the usual root, then the trees are nested, and converge locally to the infinite binary tree
. Slightly less obviously, if we take
to be one of the leaves, then the trees are still nested (up to labelling – ie in the sense of isomorphisms of rooted trees), and converge locally to the canopy tree, defined by a copy of
with nearest-neighbour edges, and where each vertex
is connected to the root of a disjoint copy of
, as shown below:
. In the case where the
are vertex-transitive, then if we only care about rooted graphs up to isomorphism, then it doesn’t matter how we choose the root.
converges in the local weak sense to
if, for all
and for all rooted graphs
,
.
, the path of length n. Then, the r-neighbourhood of a vertex is isomorphic to
, unless that vertex is within graph-distance (r-1) of one of the leaves of
. As
, the proportion of such vertices vanishes, and so,
, from which we conclude the unsurprising result that
converges in the local weak sense to
. (Which is vertex-transitive, so it doesn’t matter where we select the root.)
be the set of leaves of
, and we claim that when
is chosen uniformly from the vertices of
, then
converges in distribution. Indeed,
, whenever
, and so the given distance converges in distribution to the Geometric distribution with parameter 1/2 supported on {0,1,2,…}.
, with the choice of this vertex given by Geometric(1/2). Continue reading



